Lab 6 Rehearse 1
Hypothesis Tests Part 1
Comparing One and Two means
Lab 6 Set Up
Posit Cloud Set Up
Just as in previous labs, you will need to follow this link to set up Lab 6 in your Posit Cloud workspace:
Link to Set up Lab 6 Hypothesis Tests Part 1 Means
 Important
Important
After you have set up Lab 6 using the link above, do not use that link again as it may reset your work. The set up link loads fresh copies of the required files and they could replace the files you have worked on.
Instead use this link to go to Posit Cloud to continue to work on lab 6: Link
RStudio Desktop Setup
Link to download the Lab 6 materials to RStudio Desktop
Note that you will do all your work for this Rehearse 1 in the Lab-6-Rehearse 1 worksheet, so click on that to open it in the Editor window in your RStudio/Posit Cloud Lab 6 Hypothesis Tests 1 workspace.
Comparing One Mean to a Point Value
By “one sample - one mean” our intent is to test whether the mean of a numerical variable in a sample is different from a standard or stated value. From this we want to make an inference about the target population’s mean.
Attribution: The one-sample one mean example is based upon the explanation of the Infer Package update: Infer
Load Packages
First load the necessary packages:
 CC1
CC1
library(tidyverse)
library(infer)Inspect the Data
In this example, we are using the gss data set which is included in the Infer package. It is derived from the General Social Survey and is a sample of 500 responses to questions on the larger survey. You can find information about the GSS here.
Because the gss data set is included in Infer package which we have already loaded into the session library, we do not need to load it separately. But we need to create a data object from the gss for consistency with our process.
 CC2
CC2
gss <- gss
# Use the glimpse () function to display the data object.
glimpse(gss)## Rows: 500
## Columns: 11
## $ year <dbl> 2014, 1994, 1998, 1996, 1994, 1996, 1990, 2016, 2000, 1998, 20…
## $ age <dbl> 36, 34, 24, 42, 31, 32, 48, 36, 30, 33, 21, 30, 38, 49, 25, 56…
## $ sex <fct> male, female, male, male, male, female, female, female, female…
## $ college <fct> degree, no degree, degree, no degree, degree, no degree, no de…
## $ partyid <fct> ind, rep, ind, ind, rep, rep, dem, ind, rep, dem, dem, ind, de…
## $ hompop <dbl> 3, 4, 1, 4, 2, 4, 2, 1, 5, 2, 4, 3, 4, 4, 2, 2, 3, 2, 1, 2, 5,…
## $ hours <dbl> 50, 31, 40, 40, 40, 53, 32, 20, 40, 40, 23, 52, 38, 72, 48, 40…
## $ income <ord> $25000 or more, $20000 - 24999, $25000 or more, $25000 or more…
## $ class <fct> middle class, working class, working class, working class, mid…
## $ finrela <fct> below average, below average, below average, above average, ab…
## $ weight <dbl> 0.8960034, 1.0825000, 0.5501000, 1.0864000, 1.0825000, 1.08640…In the glimpse, we can see that the gss data object has 500 rows or observations of the variables. The variables are stored in the columns and we can see we have 11 variables.
See Glimpse for an explanation of the terms used.
Our variable of interest is the ‘hours’ variable which is quantitative. It represents the number of hours worked each week by the individuals who participated in the GSS survey.
Exploratory data analysis
Visualize the data
We will show the distribution of the data using a histogram. We have added code to calculate the mean hours worked per week and to show it on the graph with a dashed line.
 CC3
CC3
ggplot(gss, aes(x = hours)) +
geom_histogram(binwidth = 5, fill = "#69b3a2", color = "#e9ecef") +
geom_vline(aes(xintercept = mean(hours)),
color = "black", linetype = "dashed", size = 1) +
annotate("text", x = mean(gss$hours), y = 130,
label = paste("Mean:", round(mean(gss$hours), 3)),
size = 5, hjust = -.25) +
scale_x_continuous(breaks = seq(0, max(gss$hours), by = 5),
labels = seq(0, max(gss$hours), by = 5)) +
theme_light()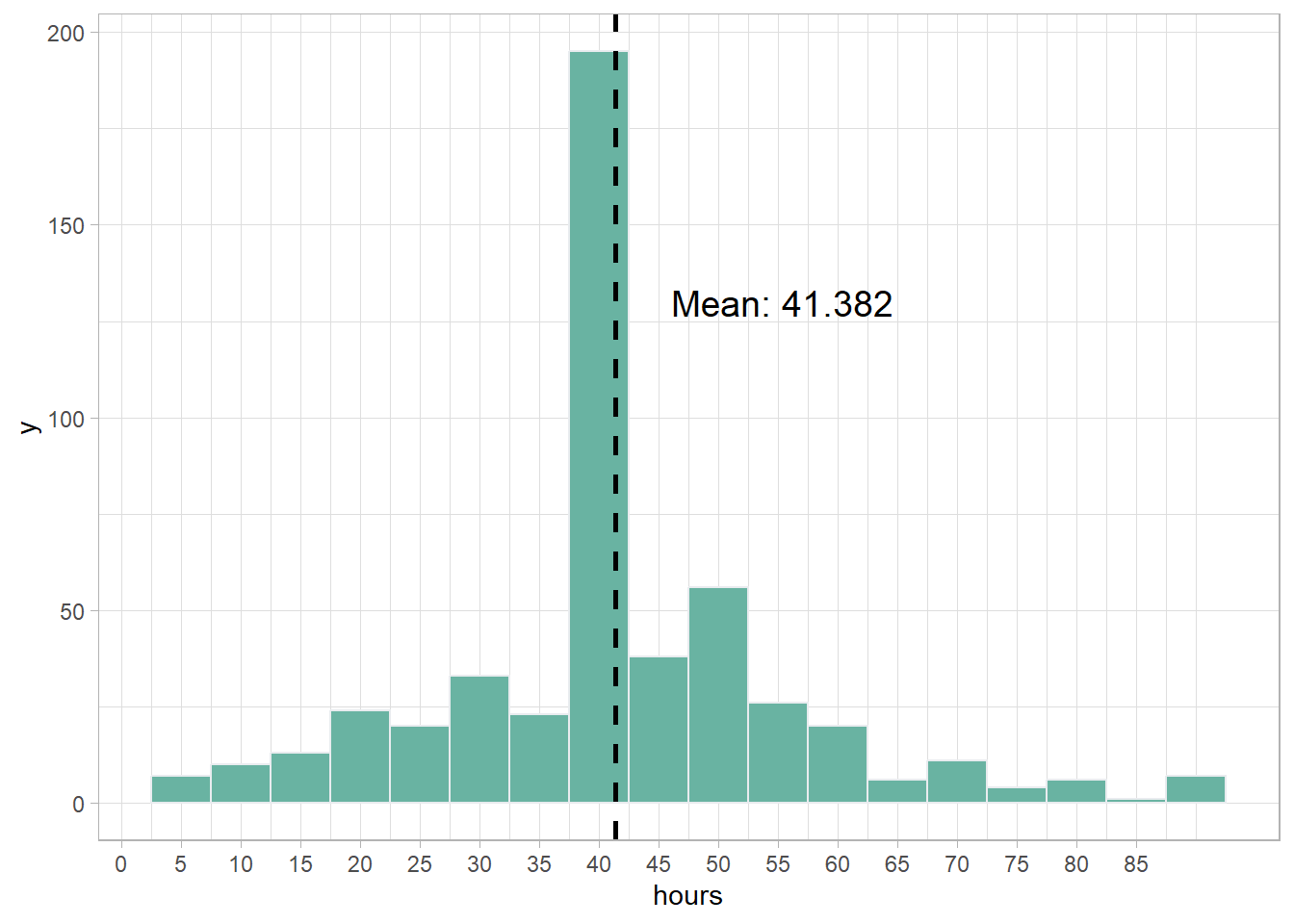
The distribution of hours in the sample has a peak near 40 hours. However, there are long tails in both directions showing a large variation in the hours worked per week by the people responding to the survey. The range is from less than 5 hours per week to more than 85 hours per week.
The mean hours worked per week in our sample is 41.382 hours per week. This seems to indicate that Americans, on average, work more than 40 hours per week.
But this is just a sample and not the entire population of American workers. So, we must take into account the variation in hours worked per week in the sample.
We do this by performing a hypothesis test on the sample data.
State the research question
Does the GSS survey provide sufficient evidence to conclude that the mean number of hours worked each week by Americans is greater than the traditional 40 hour week?
In the Null Hypothesis Significance Test (NHST), the claim, or what the researchers believe or want to prove, is stated in the Alternative Hypothesis.
The Null Hypothesis always is the exact opposite of the Alternative. Because they are exact opposites, only one can be true. If the Null is true, the Alternative must be false. If the Null is false, the Alternative must be true.
 Important
Concept: The goal of hypothesis testing is to gather evidence
against the Null in favor of the Alternative hypothesis.
Important
Concept: The goal of hypothesis testing is to gather evidence
against the Null in favor of the Alternative hypothesis.
State the Null and Alternative hypotheses
Null Hypothesis, Ho: The mean hours per week worked by Americans is less than or equal to 40.
Alternative Hypothesis, Ha: The mean hours per week worked by Americans is greater than 40.
![]()
We have stated the Null and Alternative in a “directional” format because we are interested in knowing if the mean number of hours worked per week by Americans is greater than 40. If you think of a number line, than we are interested in “proving” that mean hours worked falls to the right of 40 as it does in the histogram.
If on the other hand, we were interested in showing that Americans work fewer hours than 40 per week, the hyptotheses would be stated this way:
Null Hypothesis, Ho: The mean hours per week worked by Americans is greater than or equal to 40.
Alternative Hypothesis, Ha: The mean hours per week worked by Americans is less than 40.
This would be a “less than” or “left-tail” test.
And if we just believed that Americans do not work 40 hours per week, we would state the hypotheses this way:
Null Hypothesis, Ho: The mean hours per week worked by Americans is equal to 40.
Alternative Hypothesis, Ha: The mean hours per week worked by Americans is not equal to 40.
This would be a non-directional or “both tails” test.
 Important
Concept: The Null Hypothesis is always a form of “no
difference” or “equality.”
Important
Concept: The Null Hypothesis is always a form of “no
difference” or “equality.”
 Important
Concept: The direction of the test is determined by the
Alternative Hypothesis.
Important
Concept: The direction of the test is determined by the
Alternative Hypothesis.
Because in this example the Alternative Hypothesis contains the phrase “greater than”, this is a “right-tail” test. If we wrote the hypotheses using math symbols, the Alternative would have the > symbol. You can think of this as the > pointing to the right.
In this example we will perform the directional, right-tail test.
State the Significance Level
It’s important to set the significance level “alpha” before starting the testing using the data.
Let’s set the significance level at 5% ( i.e., 0.05) here because nothing too serious will happen if we are wrong.
Test the hypothesis - Downey Method
Here’s how we use the Downey method with
infer package to conduct this hypothesis test:
Step 1: Calculate the observed statistic
Calculate the observed statistic and store the results in the data
object called delta_obs. This is the mean of the sample
data.
![]()
Although this observed statistic is not really a difference (a
delta), we continue to use the delta_obs data object name
for consistency with our Downey/Infer process.
Because we are just interested in a single mean, we only need to
specify our response variable hours. The test statistic we
are using is the mean.
 CC4
CC4
# When reusing this code chunk in a Remix assignment:
# The `delta_obs` data object name should not be changed.
# Change `gss` to your data set name.
# change `hours` to your numerical variable.
delta_obs <- gss %>%
specify(response = hours) %>%
calculate(stat = "mean")
delta_obs## Response: hours (numeric)
## # A tibble: 1 × 1
## stat
## <dbl>
## 1 41.4Step 2: Generate the Null Distribution
Generate the null distribution of the mean hours. We use the
null_dist data object to hold this data.
We will hypothesize a Null single “point” mean value of 40. We use 40 as the Null value because the Null is correct if the mean is 40 or less.
 CC5
CC5
# When reusing this code in a Remix assignment:
# The `null_dist` data object name should not be changed.
# Change `gss` to your data set name.
# change `hours` to your numerical variable.
# Change the assumed mean value of 40 to your Null value.
# Do not change anything else.
# set a seed value for reproducible results.
set.seed(123)
# Mu stands for the assumed mean
null_dist <- gss %>%
specify(response = hours) %>%
hypothesize(null = "point", mu = 40) %>%
generate(reps = 1000) %>%
calculate(stat = "mean")
# Print first 5 rows
null_dist %>%
slice(1:5)## Response: hours (numeric)
## Null Hypothesis: point
## # A tibble: 5 × 2
## replicate stat
## <int> <dbl>
## 1 1 39.1
## 2 2 39.5
## 3 3 39.6
## 4 4 39.7
## 5 5 43.0Step 3: Visualize the Null and the Observed Statistic
Here we will plot the distribution of means in null-dist
and and the observed test statistic delta_obs.
 CC6
CC6
# When reusing this code chunk in a Remix assignment, edit only
# the axis labels,
# the title,
# and the subtitle.
# direction should be for the Alternative hypothesis which is "greater" than in this example.
visualize(null_dist) +
shade_p_value(obs_stat = delta_obs, direction = "greater")+
labs(x = "Mean Hours Worked", y = "Count",
title = "Null distribution of mean hours worked",
subtitle = "Actual mean hours worked observed (delta_obs) in the data is marked in red"
) +
theme_light()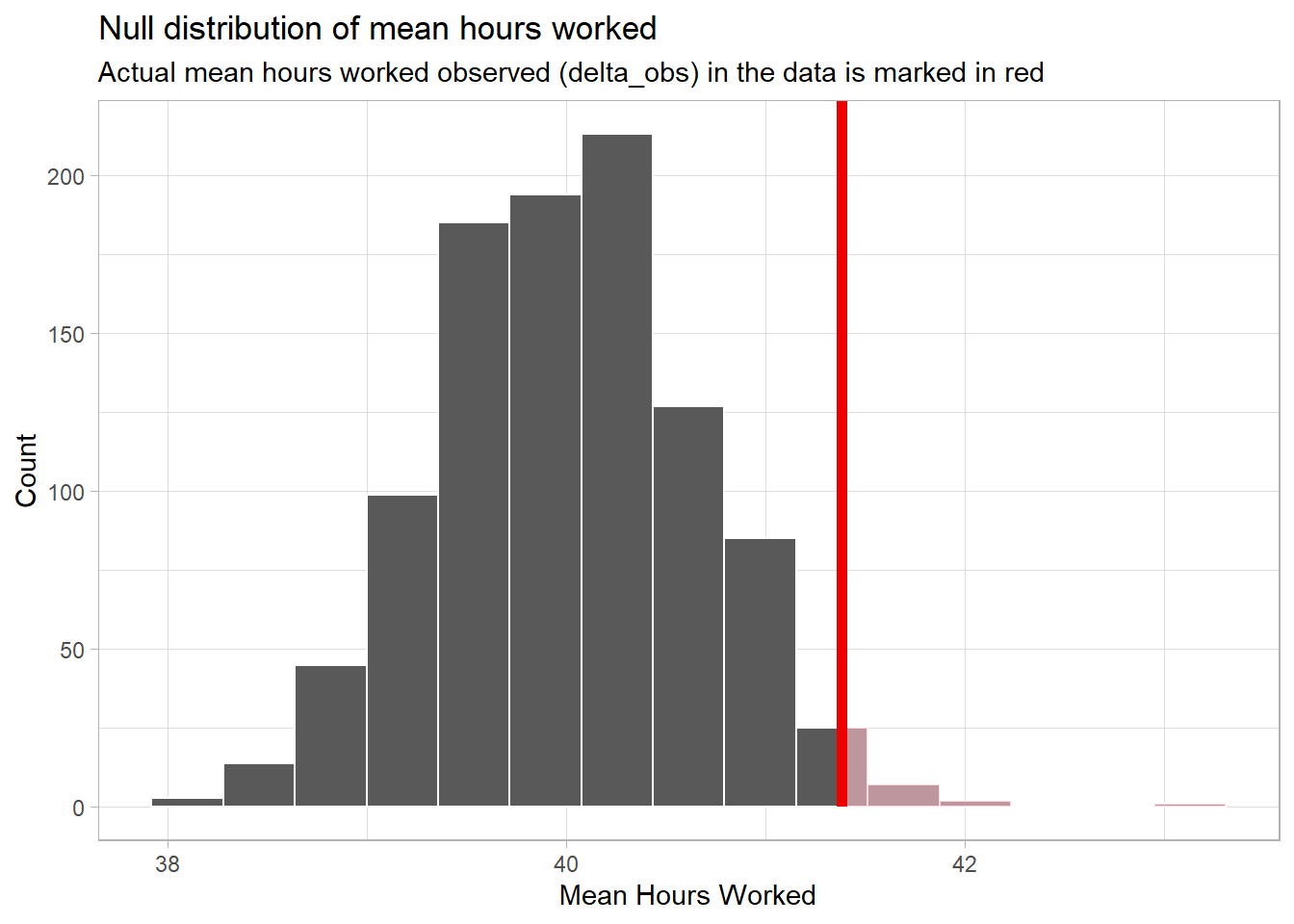

Because this is a directional, right-tail test, we include the “direction = greater” in the above code. If it were a non-directional, two-tail test, we would use “direction = both.” If it were a left-tail test, we would use “direction = less”.
The “Count” on the y-axis is the number of means in the
null_dist with a specific value of hours worked. You can
see that we do have a “bell-shaped” distribution with the center
clustered around 40 and with few observations below 38 or above 42
hours.
Notice that the “pink area under the curve” is only on the right side beyond the red line.
![]()
Question: Is the observed sample mean (delta_obs)
unlikely to happen often in the Null world? Support your answer by
explaining why you think this?
Answer here:
Step 4: Calculate the p-value
Calculate the p-value - the “area” under the curve (the distribution
of means in the Null world (in null_dist) beyond the
observed sample mean delta_obs. Again, notice this is the
pink area on the right side of the red line in the graph.
 CC7
CC7
# When reusing this code chunk in a Remix assignment, change only the direction to correspond with the alternative hypothesis. Here it is > 40, so "right" or "greater".
p_value <- null_dist %>%
get_p_value(obs_stat = delta_obs, direction = "greater")
p_value## # A tibble: 1 × 1
## p_value
## <dbl>
## 1 0.015Similar to Step 3, we set “direction =” to match the Alternative hypothesis.
Step 5: Decide if delta_obs is statistically significant
 CC8
CC8
# When reusing this code chunk in a Remix assignment:
# Replace the value for alpha with your significance level
alpha <- 0.05
if(p_value < alpha){
print("Reject the Null Hypothesis")
} else {
print("Fail to reject the Null Hypothesis")
}## [1] "Reject the Null Hypothesis"Although we illustrate making the decision on the p-value using code, you can do this with just logical thinking if you prefer. But do remember that a p-value greater than or equal to the significance level means “Fail to Reject the Null.”
Results and Conclusions
Because we set the significance level alpha to 0.05, the p-value of 0.015 is less than alpha. Thus, the Null hypothesis (mean hours worked <= 40) must be rejected and we can conclude:
The mean number of hours worked each week by Americans is greater than 40.
Calculate confidence interval
The following code chunk creates a 95th percentile confidence interval around the mean hours worked by Americans using the bootstrap method.
 CC9
CC9
# When reusing this code chunk in a Remix assignment:
# Change `gss` to your data set.
# Change `hours` to your numerical variable.
# Do not change anything else.
# Set seed to make the results reproducible.
set.seed(123)
boot_dist <- gss %>%
specify(response = hours) %>%
generate(reps = 1000, type = "bootstrap") %>%
calculate(stat = "mean")
#calculate the 95% confidence interval
ci <- get_ci(boot_dist)
# display the CI
ci## # A tibble: 1 × 2
## lower_ci upper_ci
## <dbl> <dbl>
## 1 40.1 42.6![]()
Question: State what the confidence interval tells us about the likely mean hours worked per week? Does it support the Null Hypothesis of the mean being less than or equal to 40?
Your Answer here:
Traditional hypothesis test
 CC10
CC10
# When reusing this code chunk in a Remix assignment:
# Change hours to your numerical variable..
# Change gss to your data set
# Change the Null mean mu of 40 to your value.
# Change the alternative to less for an Ha using <, or delete that section
t.test(gss$hours, mu = 40, alternative = "greater", conf.level = 0.95)##
## One Sample t-test
##
## data: gss$hours
## t = 2.0852, df = 499, p-value = 0.01878
## alternative hypothesis: true mean is greater than 40
## 95 percent confidence interval:
## 40.28981 Inf
## sample estimates:
## mean of x
## 41.382Again, we include “greater” in the code.
The results of the traditional test are similar to the Infer results. The Null of the mean hours worked being <= 40 must be rejected.
Remember there are assumptions required for the t-test that should be checked and verified before the results can be used. We will not do that here. See Assumptions for Traditional Tests
Comparing Two Means
When we need to determine whether there is a significant difference between the means of two groups, there are two types of hypothesis tests we can use: the independent samples test and the paired samples test.
Independent Samples Test: This test is used when you want to compare the means of two independent groups to determine whether there is statistical evidence that the associated population means are significantly different. The two groups are independent, meaning that they do not affect each other. For example, you might use an independent samples test to determine whether there is a difference in exam scores between students who had studied alone versus those who studied in groups.
Paired Samples Test: This test, also known as the dependent samples test, is used when you want to compare the means of the same group or item under two separate scenarios. The two scenarios are dependent, meaning that they are related or paired in some way. For example, you might use a paired samples test to determine whether there is a significant difference in a person’s heart rate before and after consuming caffeine.
In summary, the key difference between these two tests lies in the nature of the samples. Independent samples are separate and unrelated, while paired samples are related in some way. The choice of test depends on the design of your study and the nature of your data.
Independent Samples Test
In Module 5, Rehearse 2, Question 1, we used the Downey-Infer process to test for the difference in the means of two independent samples. The research question was Is there a significant difference in the mean female and male college freshman GPAs?
Note we will not repeat that information here.
Paired Samples Test
Load Packages
First load the necessary packages:
 CC11
CC11
library(tidyverse)
library(infer)Inspect the Data
Load the data.
For this example, we will be using data from research on 10 mice who
were given a treatment intended to cause them to gain weight. The data
file weights.xlsx is in the /data folder.
 CC12
CC12
# The `readxl` package is needed to load Excel worksheets.
library(readxl)
weights <- read_excel("./data/weights.xlsx")
glimpse(weights)## Rows: 20
## Columns: 4
## $ ...1 <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, …
## $ group <chr> "before", "before", "before", "before", "before", "before", "be…
## $ weight <dbl> 200.1, 190.9, 192.7, 213.0, 241.4, 196.9, 172.2, 185.5, 205.2, …
## $ diff <dbl> 12.8, 22.3, -27.6, 0.0, 12.6, 51.0, 69.8, 18.4, 7.1, -21.5, NA,…We can see in the glimpse that the data set has 20 observations of the mice in the experiment. And it has 3 variables:
groupis a categorical variable with two levels: before and after.weightis the weight (in grams) of the mice.diffis the difference between the mice weights (after - before). Note there are just 10 observations of thediffvariable.

This data set has been “wrangled” to include the “diff” variable which is needed for use of the Downey-Infer approach to paired samples problems.
State the Research Question
Is there a difference in the “before” and “after” treatment weights of the mice?
Exploratory data analysis
Visualize the data
We will use a box plot to help us visualize the data. We will use the
group variable on the x-axis and the weight
variable on the y-axis.
 CC13
CC13
ggplot(weights, aes(x = group, y = weight, fill = group)) +
geom_boxplot(alpha = 0.7) +
labs(title = "Weights before and after treatment",
x = "Group", y = "Weight(gms)") +
scale_fill_manual(values = alpha(c("#66c2a5", "red"), 0.5)) +
stat_summary(
fun = mean,
geom = "point",
shape = 18,
size = 4,
color = "yellow",
position = position_dodge(width = 0.75),
show.legend = FALSE
) +
theme_light()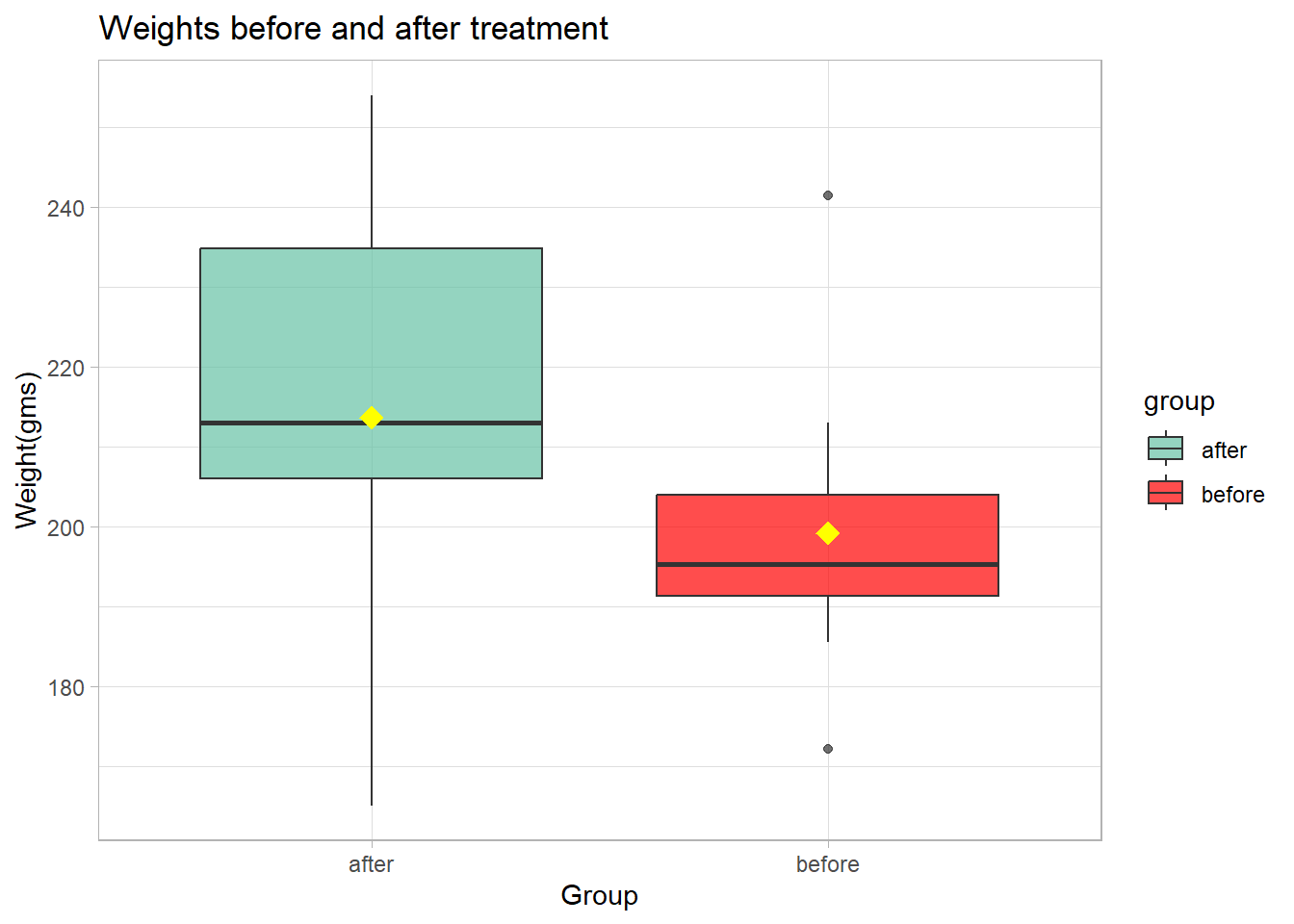
Note the standard boxplot displays the medians which are the black
lines near the “middle” of the two boxes. We can see that there is some
difference in the median weight for the two groups. The median weight
for after appears to be greater than that for
before. Our code adds yellow diamonds indicating the means
of the two groups.

The second method of visualizing the distribution below in CC14 is intended for information only. Do not use it for your Remix assignment. Use the method above in CC13!
A boxplot can tell us something about the distribution of the data points but a density plot can often tell more.
 CC14
CC14
# Calculate means for each group
mean_data <- aggregate(weight ~ group, data = weights, FUN = mean)
ggplot(weights, aes(x = weight, color = group, fill = group)) +
geom_density(alpha = 0.5) +
labs(title = "Weights before and after treatment",
x = "Weight (gms)", y = "Density") +
geom_vline(data = mean_data, aes(xintercept = weight, color = group), linetype = "dashed", size = 1) +
annotate("text", x = mean_data$weight, y = 0, label = round(mean_data$weight, 2),
vjust = -0.5, hjust = 0.5, color = "black", size = 4) +
theme_minimal()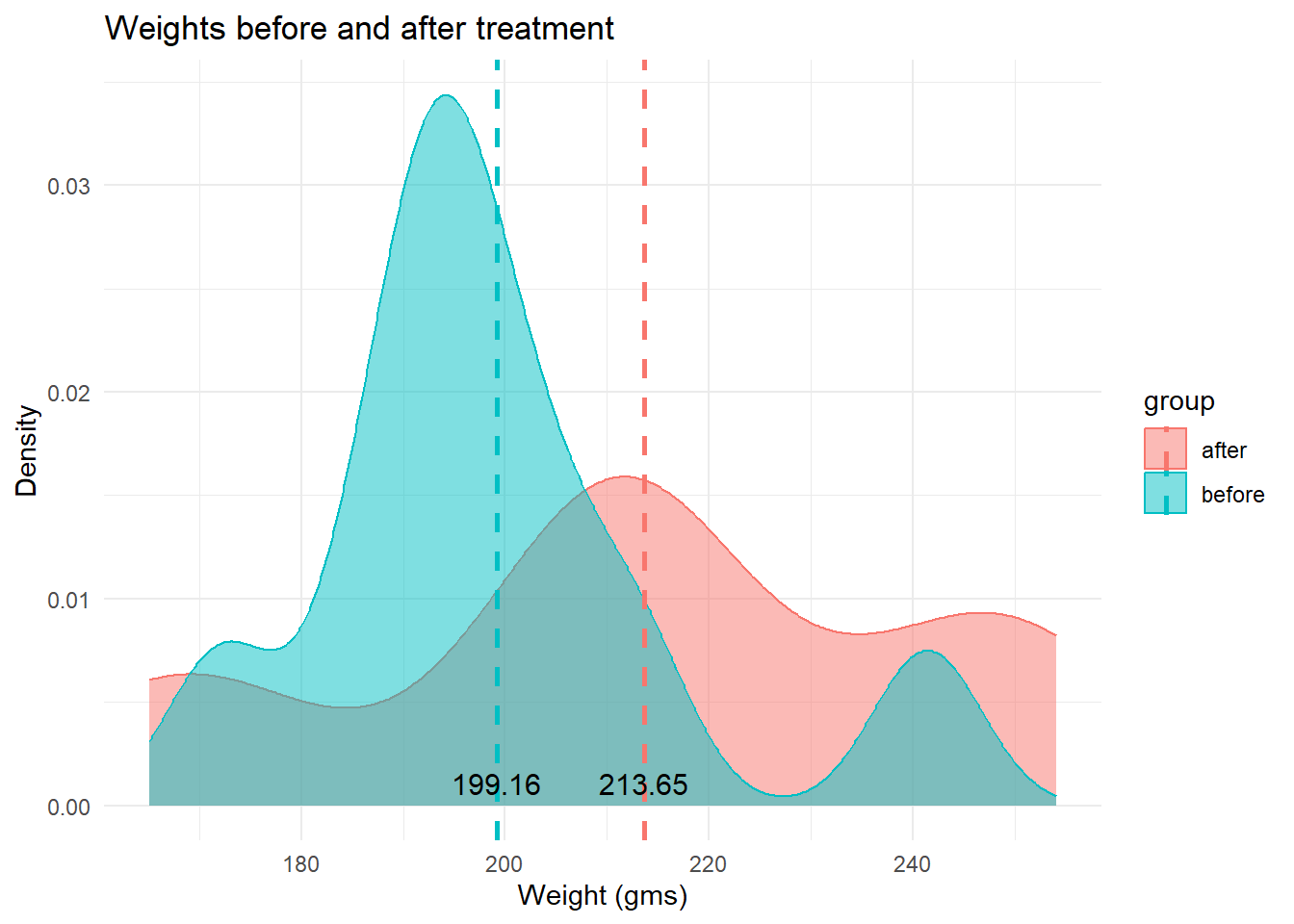
The density plot of the two distributions indicates that the
before weights are clustered around a large peak of about
195 grams, with a second, lower peak about 240 grams. The
after group has a much lower peak and is more evenly
distributed. That said, the two groups overlap each other substantially.
The mean weights of the two groups are indicated by dashed lines.
Both graphs do suggest that the mean weight of the after
group is greater than that of the before group. The mean of
the before group is 199.16 grams and that of the
after group is 213.65 grams. But is that difference enough
to be statistically significant, given the overlap in the two
distributions?
State the Null and Alternative hypotheses
Null Hypothesis Ho: There is no difference in the before treatment and after treatment weights.
Alternative Hypothesis Ha: There is a difference between the before treatment and after treatment weights.
![]()
Question: What is the direction of this hypothesis test?
Your answer here:
State the Significance Level
It’s important to set the significance level “alpha” before starting the testing using the data.
Let’s set the significance level at 5% ( i.e., 0.05) here because nothing too serious will happen if we are wrong.
Testing the hypothesis
Step 1: Calculate the observed statistic
Calculate the observed statistic and store the results in a new data
object called delta.
 Important!
Important!
The Infer package functions for conducting a paired samples test require the use of a pre-computed variable representing the calculated differences between the “before” and “after” conditions. In our course data sets, this variable is named ‘diff’. But if you use a dataset that does not have a “diff” variable, you will need to pre-compute it and add that column of values.
 CC15
CC15
# When reusing this code chunk in a Remix assignment:
# The `delta_obs` data object name should not be changed.
# Change `weights` to your data set name.
# Change `diff` to your difference variable name
# No other changes are needed.
delta_obs<- weights %>%
specify(response = diff) %>%
calculate(stat = "mean")
# Display the observed difference
delta_obs## Response: diff (numeric)
## # A tibble: 1 × 1
## stat
## <dbl>
## 1 14.5The after group has a mean weight 14.5 grams more than
the before group.
Step 2: Generate the Null Distribution
Generate the null distribution of the difference in weights.
 CC16
CC16
# When reusing this code in a Remix assignment:
# The `null_dist` data object name should not be changed.
# Change `weights` to your data set name.
# Change `diff` to your response variable.
# No other changes are needed.
set.seed(123)
null_dist <- weights %>%
specify(response = diff) %>%
hypothesize(null = "paired independence") %>%
generate(reps = 1000, type = "permute") %>%
calculate(stat = "mean")
null_dist %>%
slice(1:5)## Response: diff (numeric)
## Null Hypothesis: paired independence
## # A tibble: 5 × 2
## replicate stat
## <int> <dbl>
## 1 1 -11.6
## 2 2 11.6
## 3 3 -17.4
## 4 4 -0.970
## 5 5 18.8Note we use the hypothesize function to set our null
hypothesis to be “paired independence”. If the two variables
group and weight are independent of each
other, we should see no difference in the mean weights of the mice
before and after the treatment - the diff. We use the
calculate function to find the mean of each of the “reps” in the
null_dist data object.
Step 3: Visualize the Null and the Observed Statistic
Because this is a non-directional test, we set the “direction =” in the code to “both”.
 CC17
CC17
# When reusing this code chunk in a Remix assignment, edit only
# the axis labels,
# the title,
# and the subtitle.
visualize(null_dist) +
shade_p_value(obs_stat = delta_obs, direction = "both") +
labs(x = "Difference in Weight (grams)", y = "Count",
title = "Null distribution of Difference in Mouse Weight before and after treatment",
subtitle = "Actual difference observed in the sample data is marked in red") +
theme_light()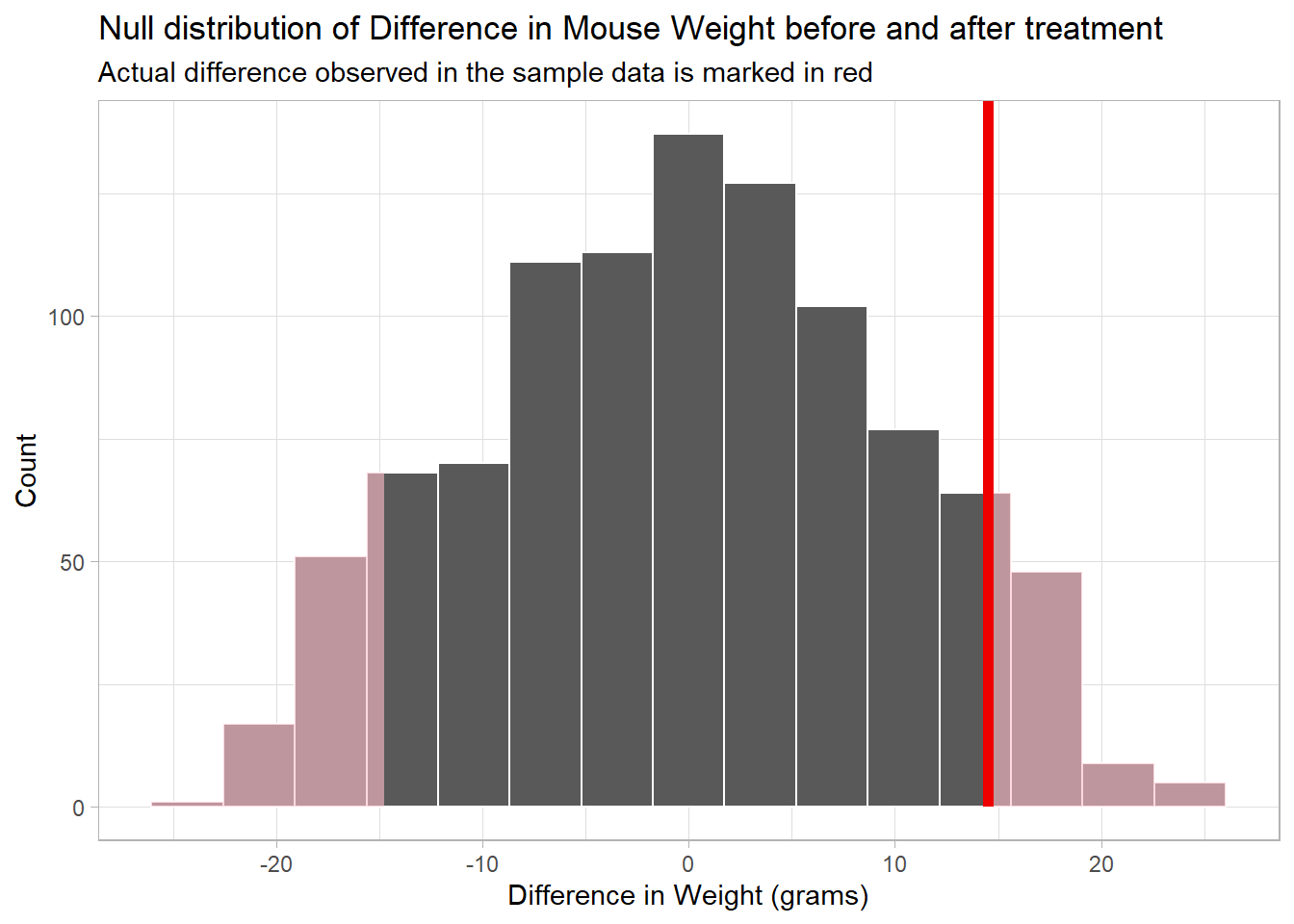

Question: Do you think the observed delta (delta_obs) occurs often in the Null world? Explain your thinking.
Your answer here:
Step 4: Calculate the p-value
Calculate the p-value (“area” under the curve beyond
delta_obs) from the Null distribution and the observed
statistic delta_obs.
![]()
We use the “both” direction because we are interested in a difference either way - a gain or a loss of weight. And that is the “pink” area under the tails in both the negative and positive directions beyond a difference of +14.9 and -14.9 grams.
 CC18
CC18
#When resuing this code chunk in a Remix assignment, no changes are needed.
p_value <- null_dist %>%
get_pvalue(obs_stat = delta_obs, direction = "both")
p_value## # A tibble: 1 × 1
## p_value
## <dbl>
## 1 0.164Step 5: Decide if delta_obs is statistically significant
 CC19
CC19
# When reusing this code chunk in Remix assignment:
# Replace the value for alpha with your significance level
# No other changes are needed.
alpha <- 0.05
if(p_value < alpha){
print("Reject the Null Hypothesis")
} else {
print("Fail to reject the Null Hypothesis")
}## [1] "Fail to reject the Null Hypothesis"Results and Conclusions
Because the p-value of approximately 0.164 is greater than the significance level of 0.05, we cannot reject the Null hypothesis of no difference.
There is no significant difference between the weights of the mice before and after the treatment.

Question: Is the finding of “no difference” surprising to you? Why or why not?
Your answer here:

Question: Do you think the substantial overlap of the distributions of the two groups (shown on the density plot) impacted whether or not the difference was signficant? Explain you thinking.
Your answer here:
Calculate confidence interval
The following code chunk will allow us to calculate a 95% confidence interval for the difference between mean mice weight “after” and “before” the treatment.
 CC20
CC20
# When reusing this code chunk in a Remix Assignment,
# Do not change the name `ci`
# Change `weights` to your data set.
# Change `diff` to your numerical difference variable.
set.seed(123)
ci <- weights %>%
specify(response = diff) %>%
generate(reps = 1000, type="bootstrap") %>%
calculate(stat = "mean") %>%
get_confidence_interval(level = 0.95)
#print the CI upper and lower limits
ci## # A tibble: 1 × 2
## lower_ci upper_ci
## <dbl> <dbl>
## 1 -2.47 32.2The Null value of 0 is in the confidence interval. This supports the conclusion that there is no significant difference in mice weight before and after the treatment.
Traditional hypothesis test
We can check the results of our Downey-Infer process by running a traditional one sample t-test for the mean difference of 0.
 CC21
CC21
t.test(weights$diff, mu = 0)##
## One Sample t-test
##
## data: weights$diff
## t = 1.5558, df = 9, p-value = 0.1542
## alternative hypothesis: true mean is not equal to 0
## 95 percent confidence interval:
## -6.578052 35.558052
## sample estimates:
## mean of x
## 14.49The traditional one-sample t-test on the diff variable
produces similar results. Because the p-value of 0.1542 is greater than
our alpha of 0.05, we cannot reject the Null of no difference in the two
means.
![]()
Question: If the two methods (Downey v traditional) produce almost identical results, why not just use the “simpler” traditional method?
Answer here:
Lab Assignment Submission
 Important
Important
When you are ready to create your final lab report, save the Lab-06-Rehearse1-Worksheet.Rmd lab file and then Knit it to PDF or Word to make a reproducible file.
Note that if you have difficulty getting the documents to Knit to either Word or PDF, and you cannot fix it, just save the completed worksheet and submit your .Rmd file for partial credit.
Submit your file in the M6.3 Lab 6 Rehearse(s): Hypothesis Tests Part 1 - Means assignment area.
The Lab 6 Hypothesis Tests Part 2 Grading Rubric will be used.

Congrats! You have finished Rehearse 1 of Lab 6. Now go to Rehearse 2!

This
work was created by Dawn Wright and is licensed under a
Creative
Commons Attribution-ShareAlike 4.0 International License.
v3.0, 2/15/26
Last Compiled 2026-02-15

